The Unstructured Orchestra: How Intrinsically Disordered Proteins and Epigenetics Challenge the Modern Synthesis
For decades, the central dogma of molecular biology and its underpinning in the Modern Synthesis have painted a clear picture: DNA makes RNA, RNA makes protein, and protein structure dictates its function. Proteins, the workhorses of the cell, were largely understood as precisely folded, rigid entities, their intricate three-dimensional shapes determining their specific roles.
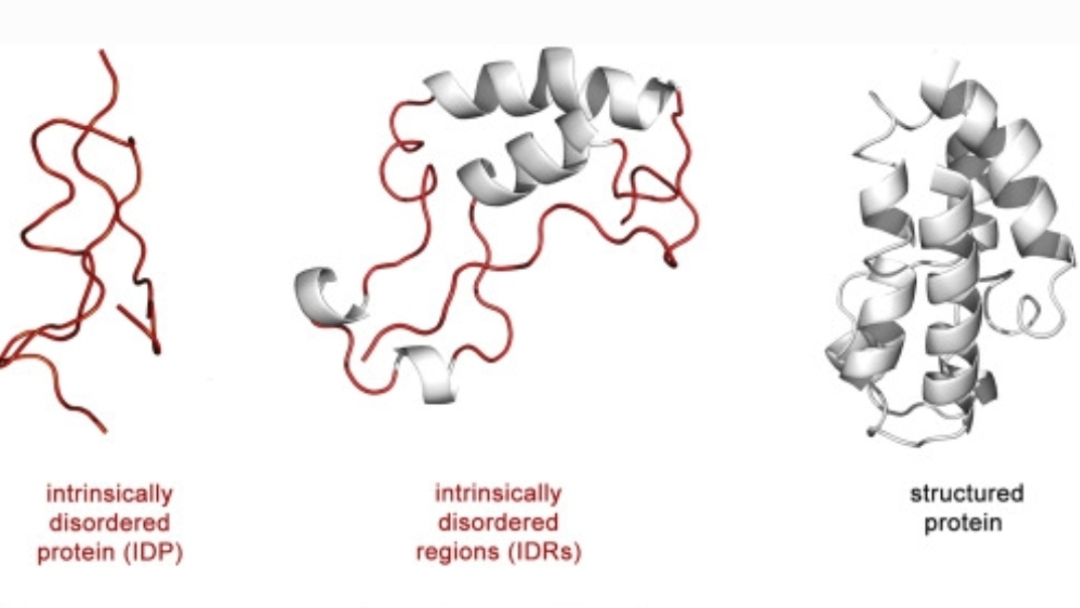
However, a growing body of evidence, particularly concerning intrinsically disordered proteins (IDPs), is forcing a re-evaluation of this paradigm, revealing a more fluid, dynamic, and indeed, messy reality within the cell. This newfound understanding not only highlights the diverse roles of IDPs in crucial cellular processes like cell signaling but also underscores the profound involvement of epigenetics, ultimately presenting a significant challenge to the traditional framework of the Modern Synthesis.Intrinsically disordered proteins, by definition, lack a stable, well-defined three-dimensional structure under physiological conditions. Far from being functionless or misfolded, these highly dynamic proteins constitute a significant portion of the proteome in eukaryotes, often exceeding 40%. Their inherent flexibility allows them to adopt a multitude of conformations, enabling them to interact with numerous binding partners in a highly promiscuous yet specific manner. This "one-to-many" and "many-to-one" binding capability is central to their diverse roles, particularly in the intricate dance of cell signaling.
In cell signaling, IDPs act as molecular rheostats, fine-tuning the amplitude and duration of signals. Their lack of fixed structure allows for rapid association and dissociation with various partners, facilitating the transient and reversible interactions critical for signal transduction pathways. For instance, many transcription factors, co-activators, and adapter proteins involved in signaling cascades are rich in disordered regions.
These regions often contain short linear motifs (SLiMs) that act as recognition sites for specific protein-protein interactions, phosphorylation events, or ubiquitination. The dynamic nature of IDPs allows them to respond to subtle changes in the cellular environment, integrating multiple signals and translating them into appropriate cellular responses, such as gene expression, cell growth, or apoptosis. Their ability to switch between different binding partners based on post-translational modifications (PTMs) is a hallmark of their regulatory power.The involvement of epigenetics in the realm of IDPs and cell signaling is profound, creating a layer of regulatory complexity that challenges the deterministic view of the Modern Synthesis. Epigenetics, the study of heritable changes in gene expression that do not involve alterations to the underlying DNA sequence, directly influences the expression, modification, and ultimately, the function of IDPs. For example, histone modifications (e.g., acetylation, methylation, phosphorylation) by enzymes like histone acetyltransferases (HATs) and histone deacetylases (HDACs) directly impact chromatin structure and accessibility.
Many of these modifying enzymes themselves contain disordered regions that facilitate their promiscuous binding to different nucleosomes. Conversely, numerous IDPs, such as components of chromatin remodeling complexes, are directly involved in "reading," "writing," and "erasing" epigenetic marks.
Consider the interplay in the context of cell signaling. An external signal might trigger a cascade that leads to the post-translational modification of an IDP, altering its conformation and binding affinity for a specific epigenetic regulator. This, in turn, could lead to changes in histone modification patterns, ultimately influencing the transcription of genes involved in the cellular response.
Conversely, the epigenetic landscape of a cell can determine the basal expression levels of certain IDPs or the enzymes that modify them, thus setting the stage for how a cell will respond to subsequent signals. This creates a feedback loop where IDP dynamics influence epigenetics, and epigenetics, in turn, modulates IDP function, adding layers of plasticity and adaptability that are difficult to reconcile with a purely gene-centric view.This intricate interplay between IDPs and epigenetics presents a significant challenge to the Modern Synthesis, which largely posits that evolution proceeds through changes in gene frequency driven by natural selection acting on heritable genetic variation.
The Modern Synthesis emphasizes a direct link between genotype and phenotype, with proteins serving as the relatively stable intermediaries. However, the prevalence and functional importance of IDPs introduce a level of phenotypic plasticity and emergent behavior not easily explained by this framework.Firstly, the "fuzziness" of IDP function, where a single protein can adopt multiple conformations and interact with numerous partners, blurs the clear-cut genotype-phenotype mapping.
The same gene sequence can give rise to a protein capable of diverse functions depending on the cellular context, PTMs, and interaction partners – many of which are themselves IDPs. This inherent flexibility suggests that evolution might also operate on the "evolvability" of protein disorder, allowing for rapid adaptation without necessarily requiring extensive genetic mutations.
Secondly, the deep involvement of epigenetics further complicates the picture. If epigenetic modifications, which are influenced by environmental cues and can be passed down through cell divisions (and sometimes even generations), significantly modulate IDP function and cellular responses, then phenotypic variation is not solely a product of genetic mutation and recombination. Non-genetic inheritance mechanisms, mediated by epigenetic changes that impact IDP behavior, introduce another layer of heritability that extends beyond the strict Mendelian inheritance patterns central to the Modern Synthesis. This suggests that adaptation and diversification can occur not just through changes in gene sequence but also through changes in gene expression and protein function mediated by environmentally responsive epigenetic mechanisms acting on dynamic IDP networks.
In conclusion, the recognition of intrinsically disordered proteins as key players in cell signaling and their profound entanglement with epigenetic mechanisms reveals a far more dynamic and adaptable cellular landscape than previously appreciated. This "unstructured orchestra" of IDPs, orchestrating cellular responses with their flexible and multi-faceted interactions, coupled with the regulatory power of epigenetics, fundamentally challenges the rigid assumptions of the Modern Synthesis. It suggests that evolution operates not just on the blueprint of genes but also on the plasticity of protein function and the heritability of epigenetic states, paving the way for a more nuanced and holistic understanding of biological complexity and adaptation.









Comments
Post a Comment